Structure Aided Drug Design
How protein structures aid in the development of drugs
Protein structures are important in the design and optimization of drugs. These drug molecules can enhance (agonist) or inhibit (antagonist) the protein’s function depending on its role in diseases. Drug design is often carried out by screening hundreds to thousands of compounds against a protein of interest and measuring its activity to determine “hits”, but this can take a long time and a lot of resources. To speed the process, understanding how a protein interacts with a drug compound can aid in the design of new drugs. One such example is a paper recently published on a novel therapeutic inhibitor for a protein called reverse transcriptase in the HIV virus.
HIV and the Goal of Developing New Inhibitors
Human immunodeficiency virus (HIV) is a chronic disease that affects over 39 million people worldwide, and if left untreated can lead to acquired immunodeficiency syndrome (AIDS). Treatment for HIV often involves a combination, or ‘cocktail’ of different drugs that target different proteins in its viral replication pathway. HIV replication involves proteins that recognize and bind to a human immune cell (CD4+ T-cell), reverse transcribe its RNA genome into double stranded DNA, and package its replicated genetic material into new viruses. One such protein that reverse transcribes the HIV RNA genome is reverse transcriptase. This protein offers a unique target for drug therapy since human cells do not have a protein that performs this function, thus limiting off-target effects if a drug is designed for the HIV protein.

There are currently FDA-approved inhibitors on the market for HIV-RT called non-nucleoside reverse transcriptase inhibitors (NNRTIs): rilpivirine (RPV), nevirapine (NVP), and doravirine (DOR). However, HIV viruses replicate so fast and without any proofreading mechanism that a lot of mutations can arise in the RNA genome. If these mutations happen to fall in the binding site for these HIV-RT NNRTIs, they will not be able to bind and reverse transcriptase will continue. Our goal was to determine a new type of NNRTI drug that maintains inhibitory activity in the face of these resistance mutations.
Cryo-EM Structure of HIV-RT with Compound 12126065
Conformational Change
The protein HIV reverse transcriptase (HIV-RT) has a known structure based on past analyses. It is composed of two domains, p66 and p51, aptly named for the number of amino acids making up each of their sequences. The p66 domain houses the active site for NNRTI inhibitors, and holds on to the extending double-stranded DNA strand as it elongates under normal functioning conditions. Thus, it’s shape has been described as a ‘hand’ with a ‘finger-palm-thumb’ shape associated with ‘grabbing’ on to a DNA strand to keep it in place during reverse transcription. When an NNRTI is bound, the HIV-RT protein undergoes a conformational change – it changes its shape as a result of compound binding. This conformational change involves moving the ‘thumb’ region of the ‘hand’ over, essentially ‘opening’ the hand to release the DNA and preventing further elongation. It was this movement that I was looking for in the cryo-EM structure, and sure enough I was able to see two-dimensional images with this movement. Comparing it to the unbound (apo) HIV-RT protein, this movement is pretty obvious. It also looks like the movement associated with rilpivirine binding, another NNRTI compound.
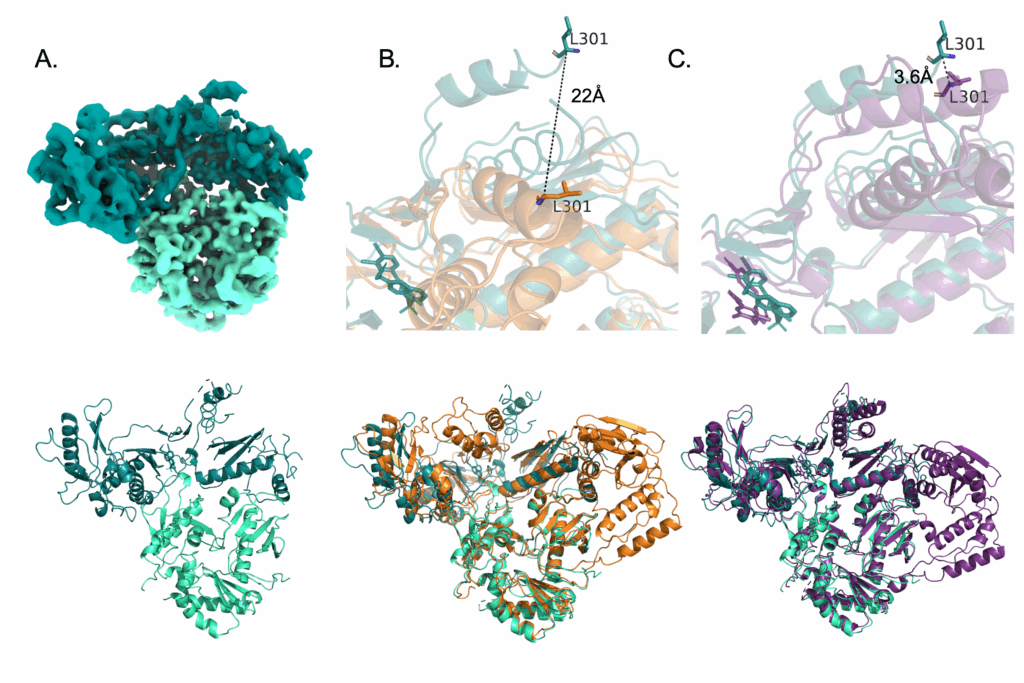
The structure solved with compound 12126065 is shown in teal. Compared to a crystal structure of HIV-RT without any bound compound (orange) you can see just how much the thumb region on the top moves (~22Å).
Compound Interactions with Protein Residues
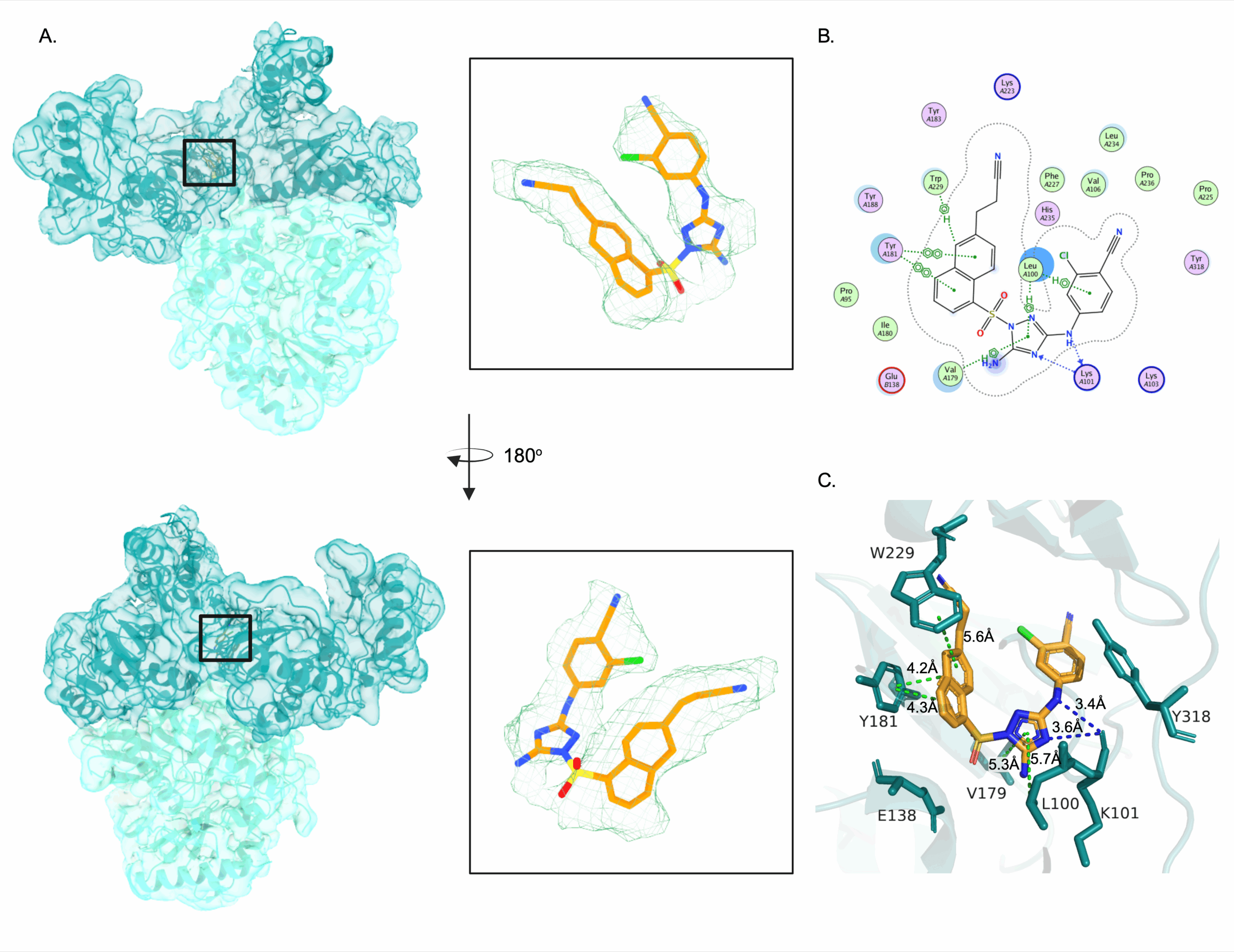
The compound we worked with has a very long name – 4-[[5-amino-1-[[6-[I-2-cyanovinyl]-1-naphthyl]sulfonyl]-1,2,4-triazol-3-ul]amino]-2-chloro-benzonitrile. This just describes the chemical composition of its structure, and for ease and clarity I’ll refer to it as compound 12126065. The binding of 12126065 to HIV-RT was in a very similar site to other NNRTIs, and the density had high resolution for placement. These interactions are the amino acid residues in the active site of the HIV-RT protein that form bonds with 12126065. Noteably, a lot of them are aromatic amino acids, with big, hydrophobic ring groups that resemble those of 12126065. The naphthalene ring of 12126065, or this double 6-membered ring region, interacts with residues Y181 and W299 for example.
When comparing 12126065 and its binding to other bound structures of NNRTIs, we see that it most resembles the structure and binding of rilpivirine (RPV). That naphthalene ring comes into play again, mainly because it is much bigger than the single 6-membered ring that RPV has in the same region. This large, bulky group could be inserting 12126065 further into the binding pocket, providing additional interactions that wouldn’t be available to RPV.

Effects of Resistance Mutations
One of the most common resistance mutations that can arise in human-infecting strains of HIV-RT are those to residues K101 and E138. Mutations to either of these residues lowers the activity of drugs RPV and NVP against HIV-RT. Taking a look at the structures of RPV and NVP in the active site of HIV-RT without mutations, we can see that these two residues form a salt bridge when the compound binds. This implies that a mutation to either of these residues would prevent formation of the salt bridge, and thus prevent binding of these two compounds altogether. However, DOR is not affected by these mutations, and in the bound structure with DOR there is no evidence of a salt bridge. Therefore, if a salt bridge is not imperative for compound binding, then mutations in these residues will not lower compound efficiency. Examining the structure of HIV-RT with 12126065, we see that there is no salt bridge between residues K101 and E138 and thus these resistance mutations will not affect this compound.

What the Structure says about Function
Identifying how compounds such as 12126065 interact with HIV-RT can lead to clues about what is critical for binding. We identified 12126065 as a binder of HIV-RT because of the conformational change induced by binding. In the active site, we identified important residues whose interactions likely aid in recognition and inhibition of reverse transcriptase activity. We also identified two resistance mutations and how their presence or absence does not affect 12126065 binding. Further improvements to the 12126065 chemical structure can now be made and tested to lower the activity of HIV-RT even more.